A beginner’s guide to metadata and ontologies
2024-07-18
Learning objectives
- define metadata and ontology in the context of biology and microbiology
- identify the importance of reliable metadata in data-centric biology
- recognize that FAIR is a process and that every step counts
Modern biology
Modern biology = big data?

[…] the real source of innovation in current biology is the attention paid to data handling and dissemination practices […] rather than the emergence of big data and associated methods per se. Leonelli (2016, 1)
Nothing is ever new
Is the attention to data handling and standardization really new in modern biology?

Model organisms
- Standardization focuses research efforts (and funds)
- But does not prohibit studying different organisms
- Quick start research on other organisms reusing findings in model organisms
Key to success: curated data description
Outline
- Why curate data?
- How to curate data?
Why curate data?
Data curation arguments
We curate data …
- in order to store for long term archive
- to give biological context for interpretation
- to be able to reanalyze in the future
- to be scrutinized against possible misconduct
(optional) Can you think of novel arguments?
Which argument is pivotal for your research?
Is is different for the research produced by others?
How to curate data?
Biological context
Biology is highly contextual (few rules, many exceptions), meaning context is the key point to address to make data travel across research investigations.
Decontextualisation
Seeing the forest for the trees
Recontextualisation
Verifying up the forest geographic coordinates
Decontextualisation

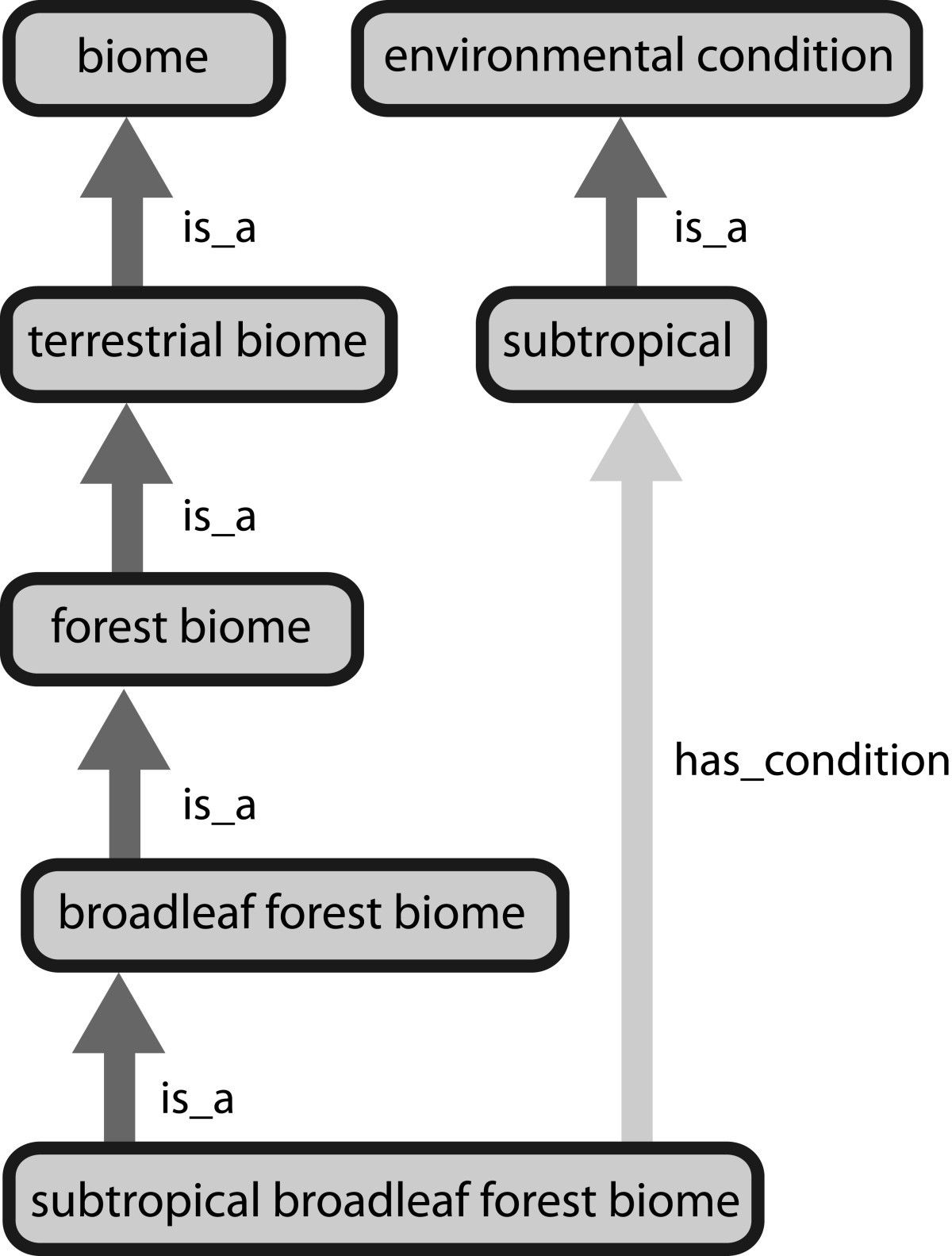
The labeling of data through bio-ontologies ensures that they are at least temporarily decoupled from information about the local features of their production. Leonelli (2016, 30)
Make data adaptable to new research settings.
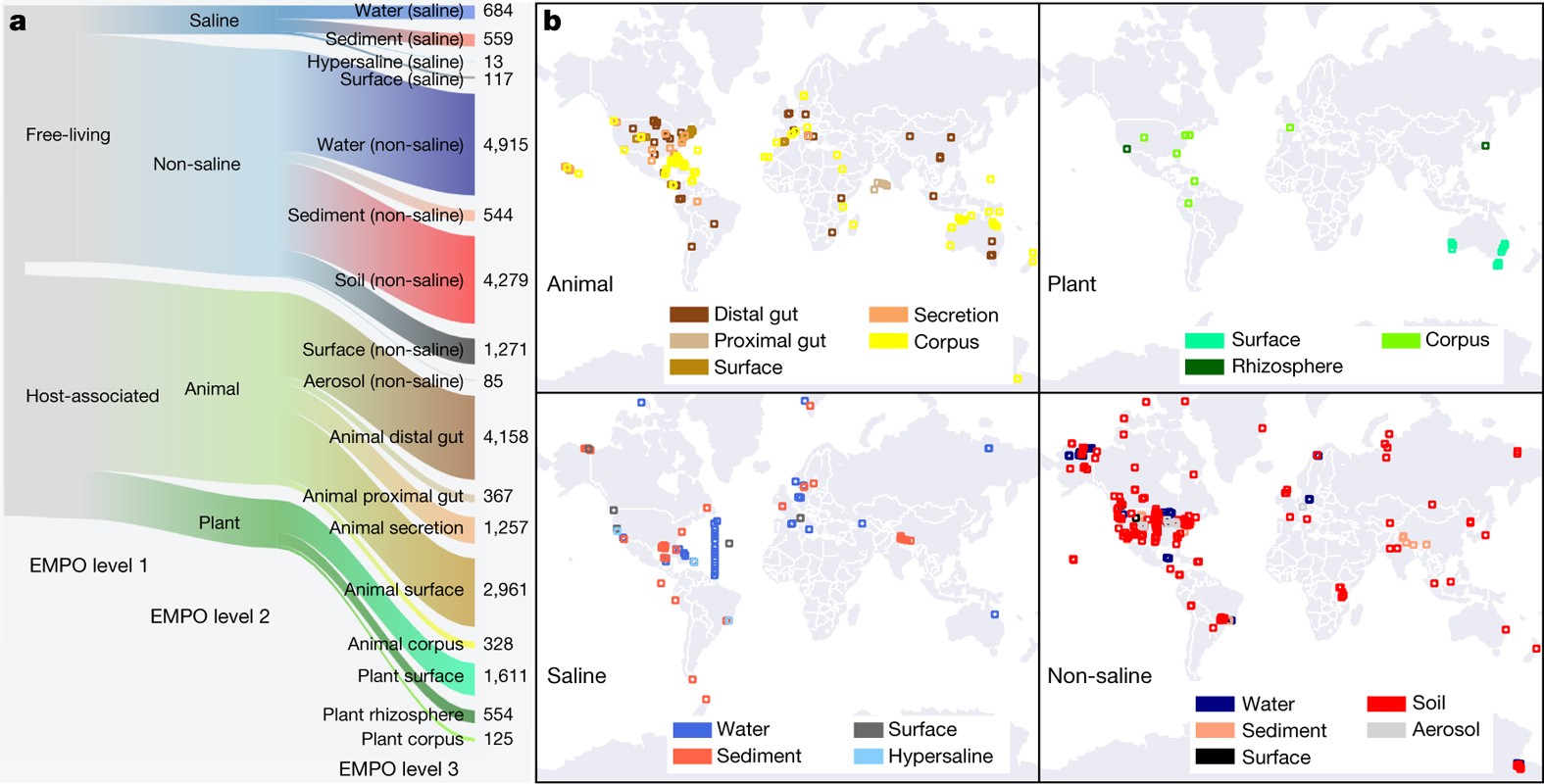
Earth Microbiome Project
100 individual studies decontextualised but recontextualised for new insights!
How to curate data?
Decontextualisation relies on ontologies
Recontextualisation?
It enables users to evaluate the potential meaning of data by assessing their provenance through the consultation of metadata. This is necessary to identify the value of data as evidence, thus helping to build an interpretation of their biological significance in a new research setting.
Leonelli (2016, 30)
Metadata definition
Metadata are data about the data, or a “love note to the future” (Scott 2011).
Metadata are “reliability labels” (Leonelli 2016, 28)
Types of metadata:
Descriptive: what is the data? e.g., title, description
Structural: how the data is organized? e.g., file, collection
Administrative: what is the provenance? e.g., versions, license
Quality: How good is the data? e.g., quality rank
Exercise: metadata
Task:
List 1-4 metadata that you have already encountered
Write them in the pad under one of the four types
In a nutshell
Forget-me-not
- Metadata are important for re-usability of your data.
- Ontologies help scientists and machines to use common terms to help generalize your data.
What about FAIR data?
FAIR principles

Source: https://scibite.com
15 principles were outlined by Wilkinson et al. (2016)
Take-home message
FAIR data takes time

FAIR data is a process
“Even if you don’t know how to go all the way to zero-to-60 open science, zero-to-20 is also really good”
Ellen Bledsoe in Perkel (2023)

References
