Biometadata Crashcourse
First steps towards FAIR (meta)data descriptions, ontology terms and data deposition
2024-12-04
Thank you for joining!
Biometadata Crashcourse: First steps towards FAIR (meta)data descriptions, ontology terms and data deposition
About us
Lectures created by
- Charlie Pauvert
- Maja Magel
- input from helpers & participants of previous Biometadata Workshops
Info
- first attempt at very short workshop version
- alternating information about concepts and hands-on exercizes
Code of Conduct
From the The Carpentries Code of conduct:
- Use welcoming and inclusive language
- Be respectful of different viewpoints and experiences
- Gracefully accept constructive criticism
- Focus on what is best for the community
- Show courtesy and respect towards other community members
You will learn
- how to prepare the necessary metadata according to FAIR principles to ease the upload of your data to a public repository of your choice
- how to identify and apply minimal metadata standards
- FAIR descriptions of your datasets – Ontology terms and how to use them
What are your expectations?
What is your background with data deposition and what are your expectations? Note them in the pad.
A beginner’s guide to metadata and ontologies
Modern biology = big data?

[…] the real source of innovation in current biology is the attention paid to data handling and dissemination practices […] rather than the emergence of big data and associated methods per se. Leonelli (2016, 1)
Nothing is ever new
Is the attention to data handling and standardization really new in modern biology?

Model organisms
- Standardization focuses research efforts (and funds)
- But does not prohibit studying different organisms
- Quick start research on other organisms reusing findings in model organisms
Key to reusability success: curated data description and published (meta)data
Success story
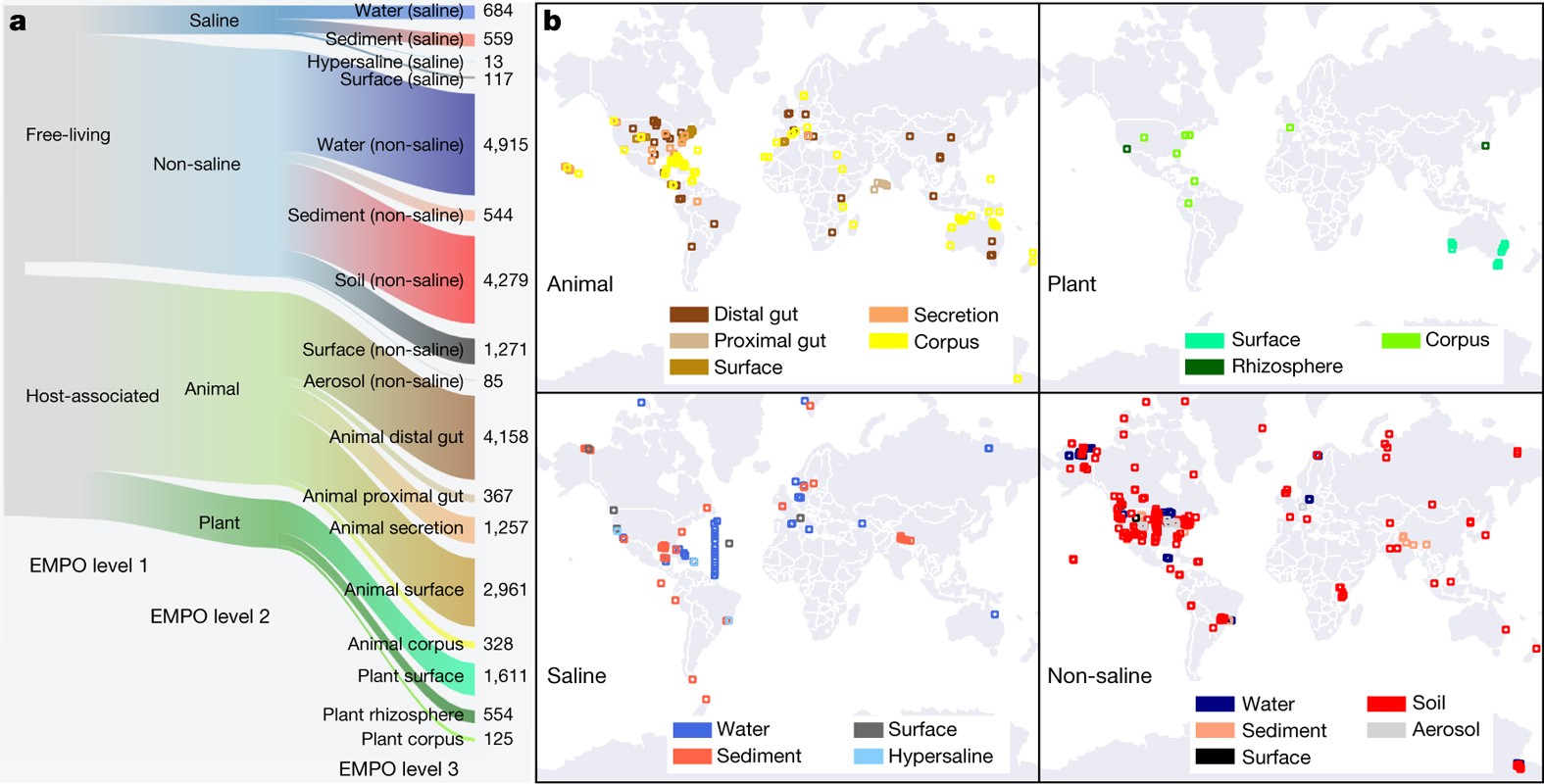
Earth Microbiome Project
100 individual studies decontextualised but recontextualised for new insights!
1. Where should you publish your data?
Data repositories
Curated and well-described that stays on your hard drive is of limited interest to the scientific community.
National and international efforts exist to create and maintain data repositories for the life sciences.
For nucleotide sequence data, the INSDC integrate and mirrors data repositories from three regions:
USA with the NCBI
Europe with the EMBL-EBI
Japan with the DDBJ
Exercise: Data repositories
Task
- Go to https://www.ebi.ac.uk/submission
- Find out which repository could suit your data using the wizard
Report the repository in the pad
homework: Where do your peers usually publish their data?
alternatively follow NFDI4Microbiota’s concise cheatsheet

2. Depositing data means adding descriptive metadata
Metadata definition
Metadata are data about the data, or a “love note to the future” (Scott 2011).
Metadata are “reliability labels” (Leonelli 2016, 28)
Types of metadata:
Descriptive: what is the data? e.g., title, description
Structural: how the data is organized? e.g., file, collection
Administrative: what is the provenance? e.g., versions, license
Quality: How good is the data? e.g., quality rank
Exercise: metadata
Task:
List 1-4 metadata that you have already encountered
Write them in the pad under one of the four types
Let’s choose ENA and figure out our required metadata
Metadata fields
- A column header expects one or more cell values
- A metadata field expects one or more values
| Metadata field | Type of constraint |
|---|---|
| description | free-text |
| geolocation | coordinates |
| biome | ontology term |
Metadata standards
Fields are organized into coherent metadata standard for a given type of data
- e.g., genomes, soil samples
They are built and maintained by a combination of stakeholders
- e.g., users community, data repositories
Standards expectations
Metadata standards (should) indicate for each field:
the description of the metadata field
the level of requirements (mandatory, recommended, optional)
the cardinality, that is the range of expected values for the metadata field
a persistent identifier for the field
Exercise: Metadata standards
Task:
Look-up a standard that match your model system or type of sample
Identify the mandatory metadata fields
Compare with your neighbor
Minimal requirements
The set of mandatory fields is sometimes referred to as the minimal requirements.
Filling out these requirements and all the optional metadata fields would be ideal (if only possible) but is time-consuming
NFDI4Microbiota started to collect overlap of minimal metadata fields across different platforms, data types and biological & environmental metadata
Exercise: Requirements
Task:
- Given only the mandatory fields, do you think you could recontextualise the data properly?
- List your arguments in the pad & note down necessary metadata fields to enhance the understanding for your dataset
ENA Browser for host-associated metadata requirements (ERC00013)
Reusability requires context & metadata
Biological context
Biology is highly contextual (few rules, many exceptions), meaning context is the key point to address to make data travel across research investigations.
Why should you share this information?
Rationale
In space, no one can hear you scream.
In data repositories, no one can hear your data description. Take the time to describe your data for humans and machines to understand!
Illustration
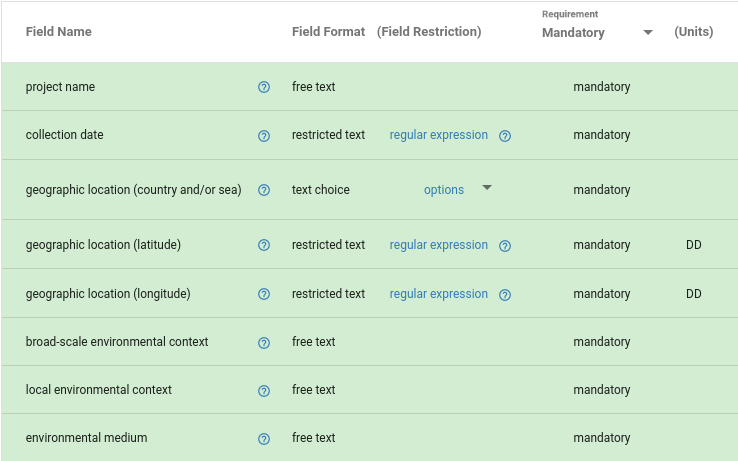
Environmental metadata according to established metadata standards
MIxS lists three mandatory environmental metadata fields that expect ontology terms.
| Metadata field | Abbreviation | Definition |
|---|---|---|
| broad-scale environmental context | env_broad_scale |
global correlation; ecosystem |
| local environmental context | env_local_scale |
in local vicinity; causal influences |
| environmental medium | env_medium |
immediate surroundings of your sample during sampling |
| Metadata field | Abbreviation | Recommended use of subclasses from |
|---|---|---|
| broad-scale environmental context | env_broad_scale |
biome [ENVO:00000428] |
| local environmental context | env_local_scale |
deeper hierarchy than broad-scale (UBERON terms accepted) |
| environmental medium | env_medium |
environmental material [ENVO:00010483] |
3. Describing human and machine-readable data
Focusing on biological and environmental metadata
What can you tell us about the biological & environmental provenance of your dataset or sample?
Where are your samples from? Do you use a model organism?
How can you make this information machine-accessible?
Why bother with ontologies?
- increase findability of your dataset
- improve machine-readability of your datasets
- help others correctly categorize & re-use your datasets > recontextualization
- required by data repositories
- first step towards open linked data and knowledge graph representations
Having fun with ontologies
Let’s talk about…
Having fun with ontologies


- Were you thinking of the baked goods, such as
cake food product [FOODON:00001278]? - unfortunately there was no well described ontology term for
pie - Or were you thinking of the
pie [44315]gene that encodes thepineapple eye protein (fruit fly) [PR:Q9VKW2]?
Ontology definition
List of terms, usually taken from the scientific literature
Ontology terms:
have curated textual definitions and synonyms
are arranged in a hierarchy from general to specific
have defined relationships with others terms (e.g.,
is_a,has_condition)have persistent identifiers
can be cross-referenced with other resources (ontology or not)
Ontology should reflect existing knowledge
Search terms in ontologies
You can find terms in ontologies using the search bar of:

- others: The OBO Foundry, OntoBee mainly for developers
Ontology Lookup Service
OLS is the official ontology service of the EMBL-EBI
- EMBL: European Molecular Biology Laboratory
- EMBL-EBI: EMBL’s European Bioinformatics Institute
Demonstration: navigating OLS for term “intestines” in UBERON, ENVO
Ontology definition as demonstrated with the term “intestine”
Results, we found the term
intestine [UBERON:0000160]
in both ontologies UBERON and ENVO (as imported term from UBERON)
The correct writing convention for ontology terms and their term identifiers is: > term [ontology-acronym:sequence-number], e.g. intestine [UBERON:0000160]
Exercise: Ontology browser
Task: using Ontology Lookup Service v4
Look-up the following keywords in that order via the search bar: pond, ear and leaf
Select a term for each using these ontologies:
Uber-anatomy ontology (UBERON)
Plant Ontology (PO)
Environmental Ontology (ENVO)
Report the term and the term identifier in the pad

Exercise: Find your ontology terms & connect the dots
Can we start the next lesson on selecting ontology terms for YOUR dataset descriptions?
TASK: Pick any 3 terms from your dataset description.
- Note them in the pad.
- Browse the OLS and find 1-2 suitable ontology terms for each.
- Add the ontology term identifiers to the pad.
- You have 5 minutes.
- Share your experiences.
- Who used an ontology besides UBERON, ENVO and PO? And why?
Reminder: Selective ontology terms for env & biological metadata
MIxS lists three mandatory environmental metadata fields that expect ontology terms.
| Metadata field | Abbreviation | Recommended use of subclasses from |
|---|---|---|
| broad-scale environmental context | env_broad_scale |
biome [ENVO:00000428] |
| local environmental context | env_local_scale |
deeper hierarchy than broad-scale (UBERON terms accepted) |
| environmental medium | env_medium |
environmental material [ENVO:00010483] |
Exercise: Env* metadata

Task alone or by pairs:
Browse ENVO or UBERON (see previous table)
List ontology terms fitting your data
Fill out the following template on the pad:
env_broad_scaleenv_local_scaleenv_medium
trouble finding an appropriate term downstream of the recommended class? See instructions of using other ontologies with the MIxS standard
Consolidating your metadata with DataHarmonizer
Data and metadata collection can be crucial (e.g., COVID-19)
DataHarmonizer: a super spreadsheet to help (Gill et al. 2023)
Main features
Load metadata standards
Fill the template
Validate your metadata against the template
NMDC submission portal
- National (USA) Microbiome Data Collaborative
Leverage DataHarmonizer to lower barriers to collect, study and biosample data
Not going to use it for data submission but data description!
Receive guidance on how to meet standards
Submission portal Demo
An otter dataset
- Sample: feces collected in the wild
- Model system: Eurasian river otter (Lutra lutra)
See the “full” methods section in the pad!

Exercise/demonstration
Task:
- Go to https://data-sandbox.microbiomedata.org/submission/
- Log with your ORCID and go again to the link above
- Create a new submission using the otter dataset
- Read the methods section in the pad
- Download the XLSX template to help you validate your submission
your turn!
In a nutshell
Forget-me-not
- Metadata are important for re-usability of your data.
- Ontologies help scientists and machines to use common terms to help generalize your data.
Take-home message
as much as possible, as little as necessary
Working FAIRly takes time and effort1
How much metadata is necessary to understand your research and to enable (inter-) disciplinary research?
FAIR data takes time

FAIR data is a process
“Even if you don’t know how to go all the way to zero-to-60 open science, zero-to-20 is also really good”
Ellen Bledsoe in Perkel (2023)

References